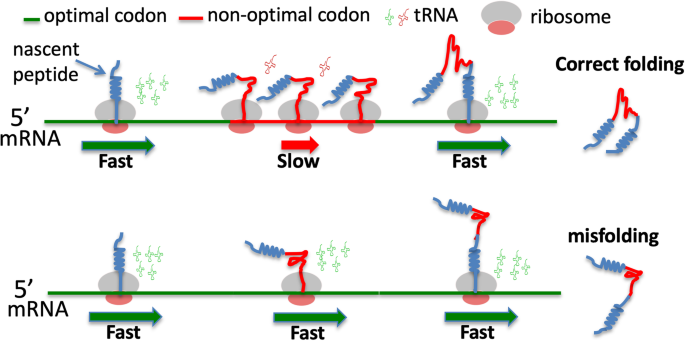
A code within the genetic code: codon usage regulates co-translational protein folding | Cell Communication and Signaling | Full Text
Comparative Analysis of Codon Usage Bias and Codon Context Patterns between Dipteran and Hymenopteran Sequenced Genomes | PLOS ONE
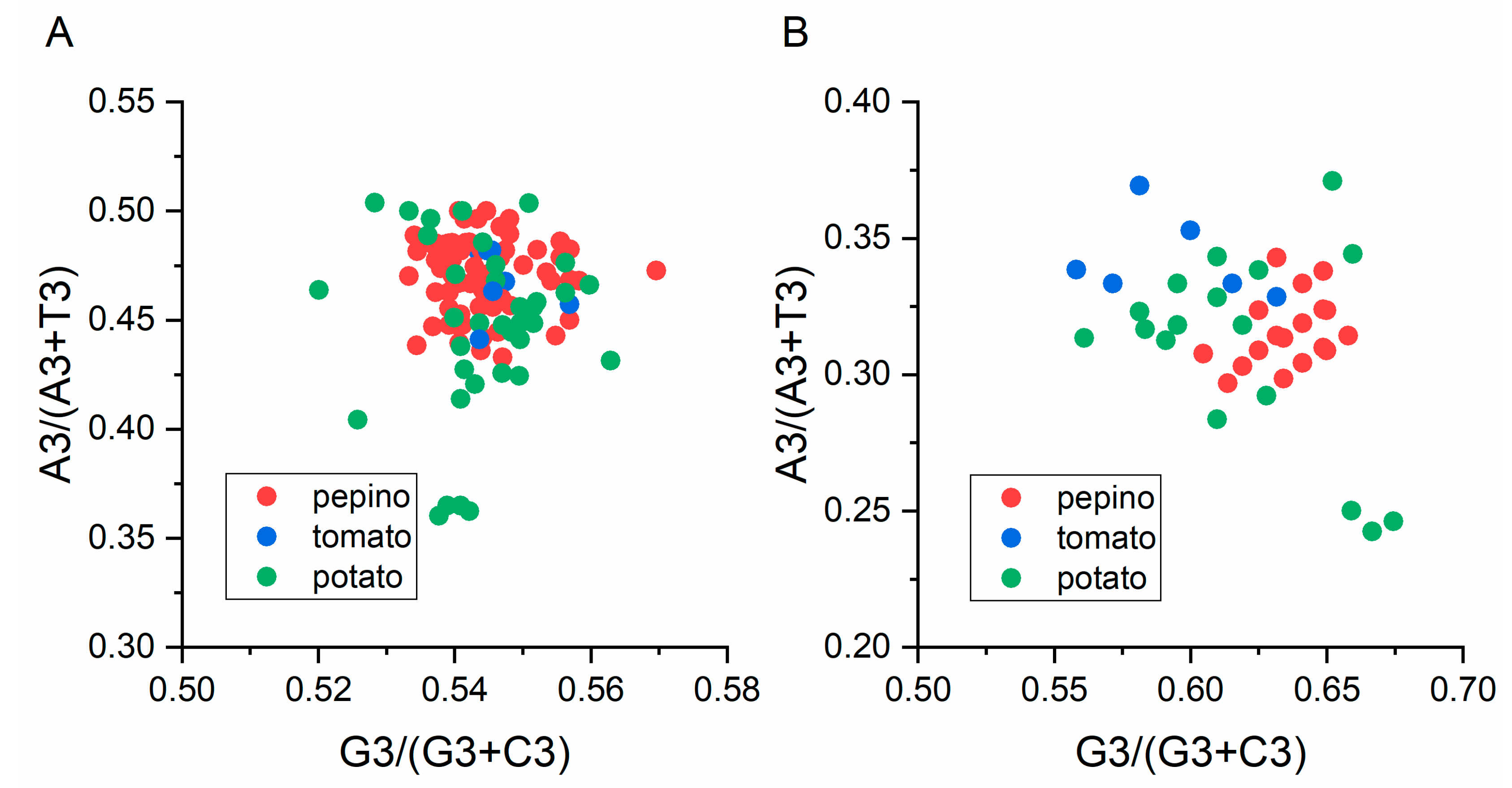
Viruses | Free Full-Text | Analysis of Synonymous Codon Usage Bias in Potato Virus M and Its Adaption to Hosts
Calculating and comparing codon usage values in rare disease genes highlights codon clustering with disease-and tissue- specific hierarchy | PLOS ONE
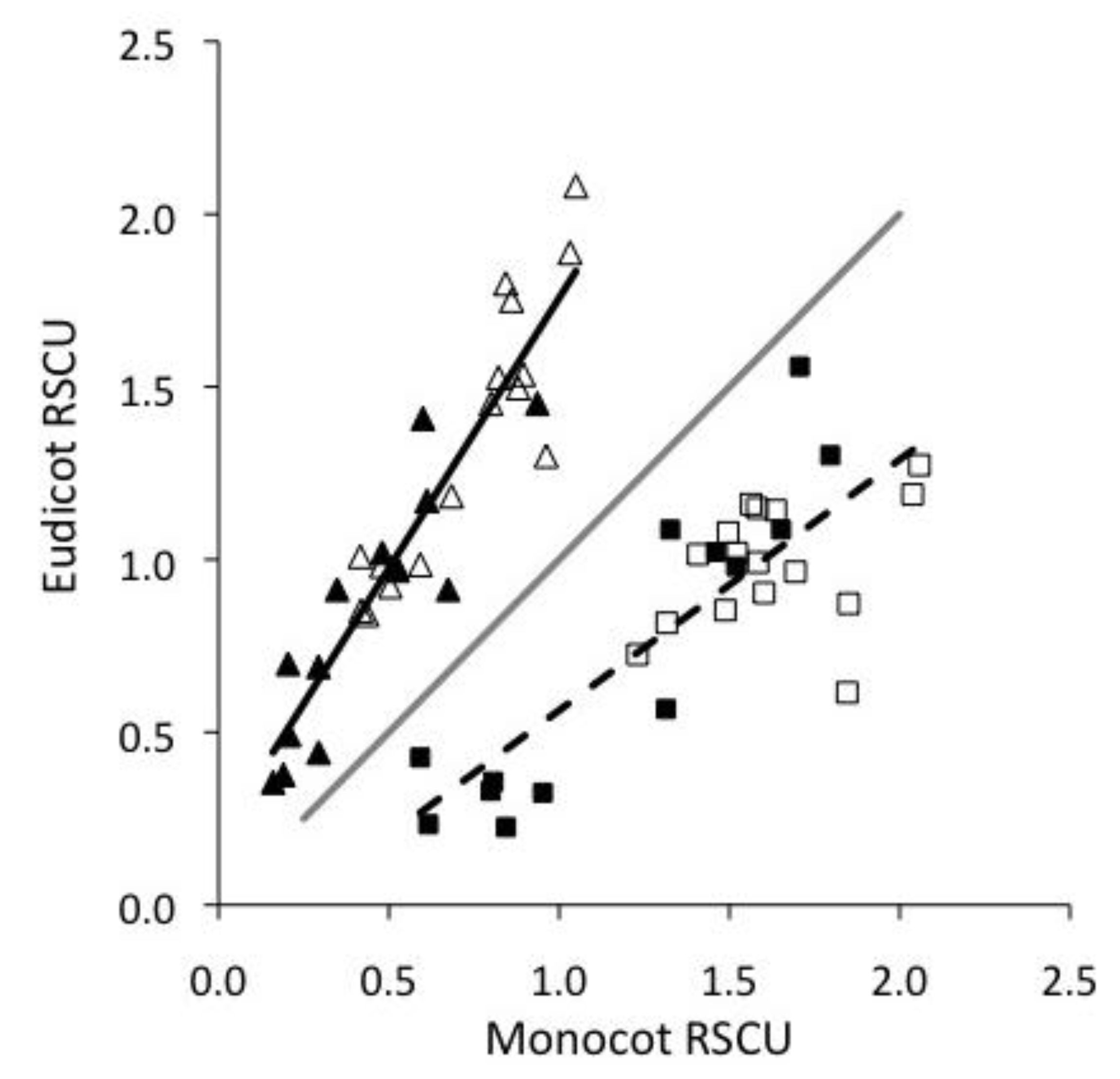
Viruses | Free Full-Text | Base Composition and Translational Selection are Insufficient to Explain Codon Usage Bias in Plant Viruses

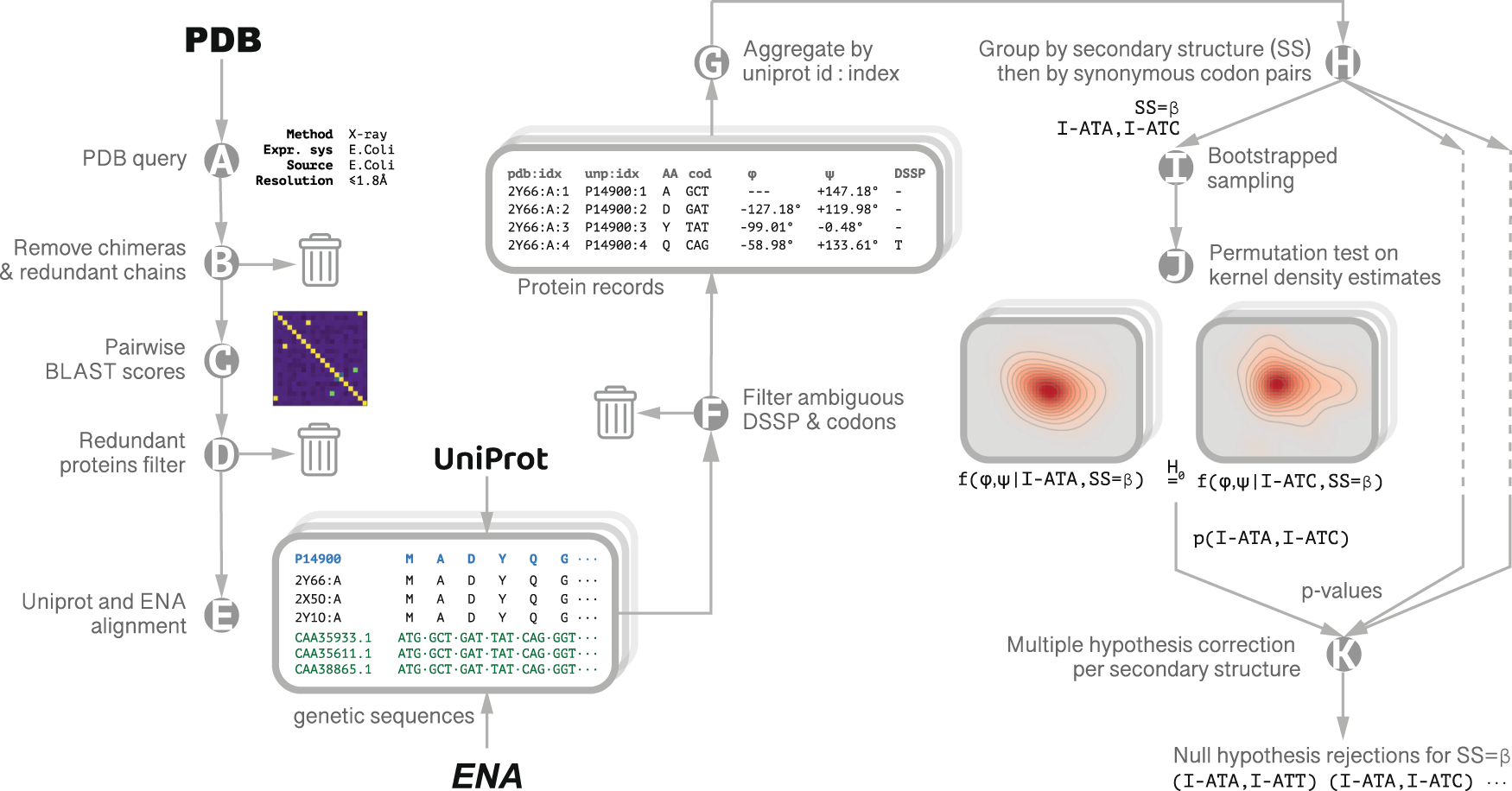
MinMax: A versatile tool for calculating and comparing synonymous codon usage and its impact on protein folding - Rodriguez - 2018 - Protein Science - Wiley Online Library

Differences in codon bias and GC content contribute to the balanced expression of TLR7 and TLR9 | PNAS
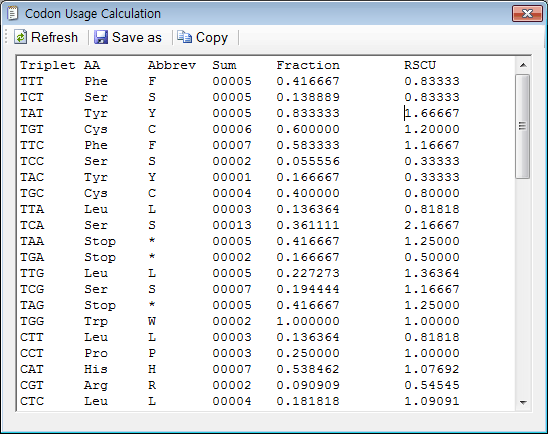
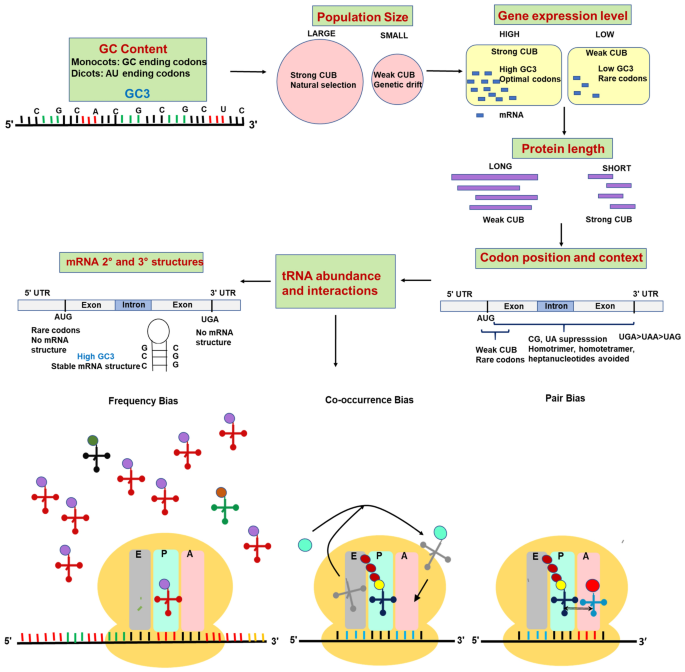
![Analysis of synonymous codon usage patterns in sixty-four different bivalve species [PeerJ] Analysis of synonymous codon usage patterns in sixty-four different bivalve species [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2015/1520/1/fig-3-full.png)



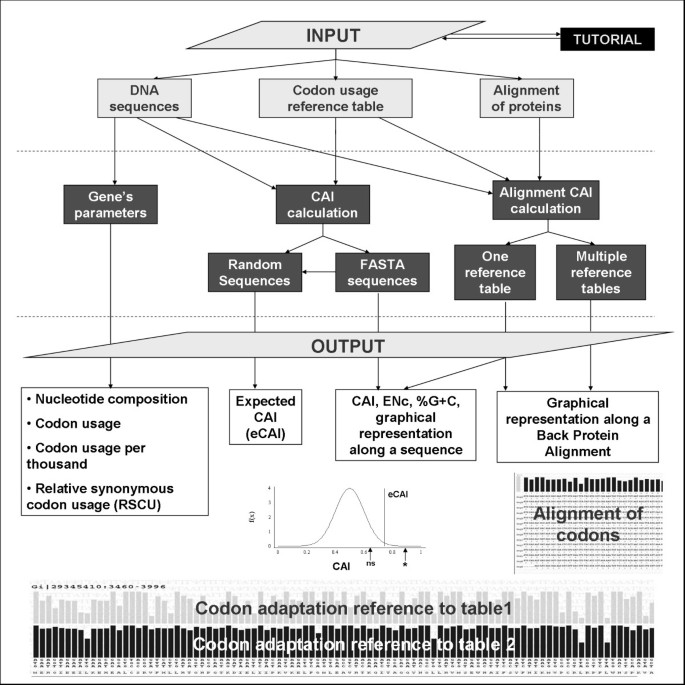
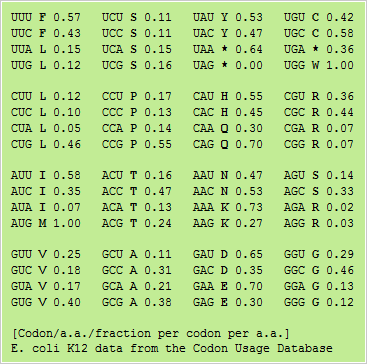

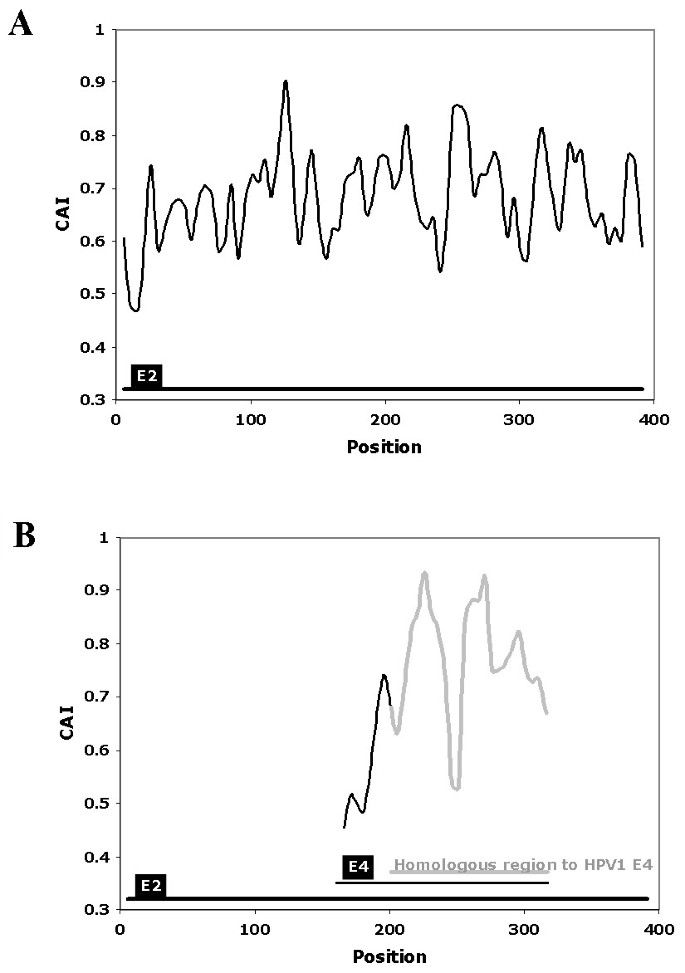
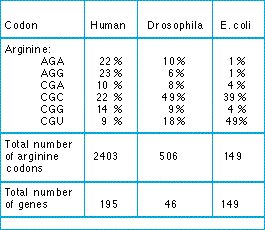
![Analysis of synonymous codon usage patterns in sixty-four different bivalve species [PeerJ] Analysis of synonymous codon usage patterns in sixty-four different bivalve species [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2015/1520/1/fig-2-2x.jpg)

